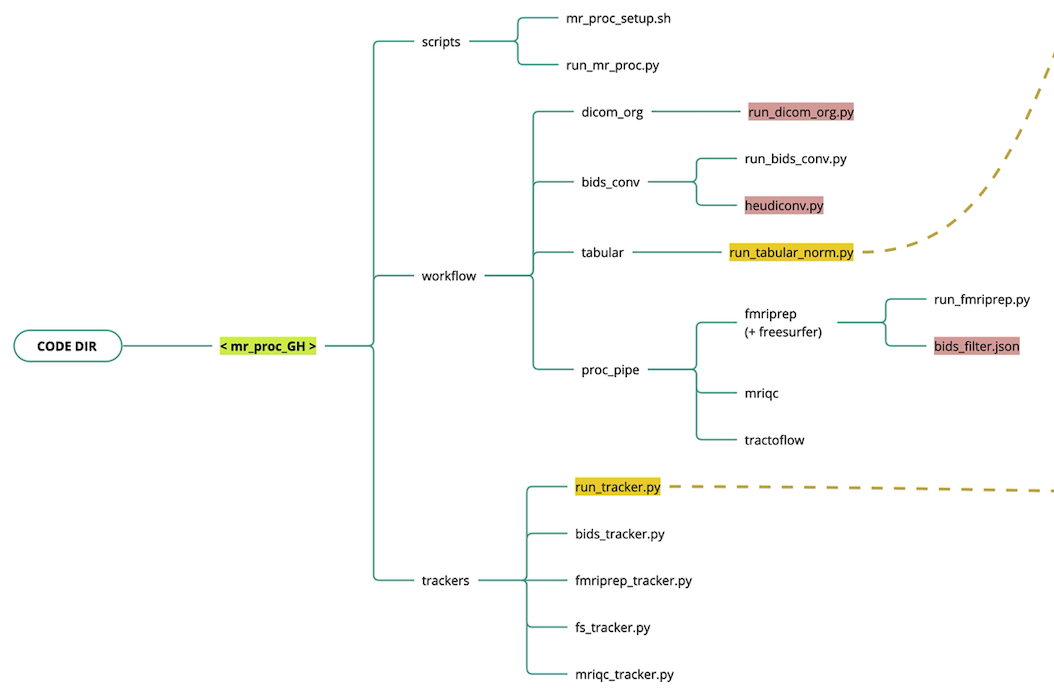
Code organization
Code organization
The Nipoppy codebase is divided into data processing workflows and data availability trackers.
workflow
- MRI data organization (
dicom_organdbids_conv)- Custom script to organize raw DICOMs (i.e. scanner output) into a flat participant-level directory.
- Convert DICOMs into BIDS using Heudiconv
- MRI data processing (
proc_pipe)- Runs a set of containerized MRI image processing pipelines
- Tabular data (
tabular)- Custom scripts to organize raw tabular data (e.g. clinial assessments)
- Custom scripts to normalize and standardize data and metadata for downstream harmonization (see NeuroBagel)
trackers
- Track available raw, standardized, and processed data
- Generate
bagelsfor Neurobagel graph and dashboard.
Legend - Red: dataset-specific code and configuration files - Yellow: Neurobagel interface